A new application profile for Preclinical Imaging has recently become available in Coscine. In this blog post, you will learn what this profile can do, how it was created, and how it can be used.
CRC 1382 Application Profile
Within the Collaborative Research Center 1382 Gut-Liver Axis, researchers and data stewards have collaborated on an application profile that can be used specifically for the area of preclinical imaging. This application profile is unique in that it is linked to three other application profiles to target imaging data for the research question.
The overarching Preclinical Imaging is connected to separate AP’s titled MRI Imaging, CT Imaging, and Organ Segmentation. Researchers can add multiple entries for a specific animal used in their experiments, specifying the different tests performed and the organs involved. This metadata is stored with the imaging files in Coscine.
Automating the Coscine Process
As application profiles are tailor made to meet the data management needs of the researcher the workflow is always at the forefront of the data steward’s mind. The goal is always to manage data that adheres to the FAIR principles while not adding unnecessary obstacles or extra steps for the researcher. For this reason, a Python library is being developed and provided by the Coscine community to enable efficient communication to and from Coscine. This makes it possible to automatically upload files to Coscine, while also extracting metadata and populating the required metadata fields in the application profile with the appropriate information.
Example
An application profile already exists, which is used for FACS files.. FACS files, or FCS files, contain raw data from flow cytometry experiments. Flow cytometry is a technique used in biology and medicine to analyze and quantify the physical and chemical characteristics of cells or particles in a fluid sample. Fortunately, FACS files follow a standard file format, facilitating compatibility and analysis using dedicated software programs.
While in the first term of SFB 1382 an application profile for FACS was developed together with researchers, now in the second funding period (starting July 1, 2023) the focus is on an efficient connection to Coscine, which will be done by a dedicated Python script.
However, to ensure that this can be used by researchers who do not have Python skills, work is underway to create a user-friendly website that will automatically run the code in the background and load the files and metadata into Coscine. This website represents an intermediate step to process a specific type of data. In this case, metadata is extracted from FACS files and transferred to a selected resource along with the files. The website is initially released to the group of researchers working with FACS data.
To use the functions of the website, it is first checked whether the researchers are authorized users of Coscine. This is done by entering the personal Coscine token.
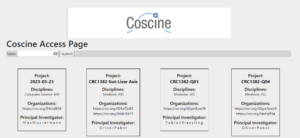
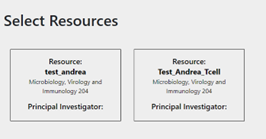
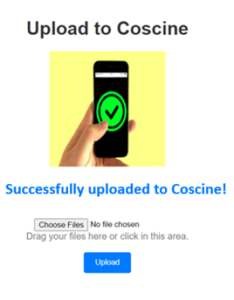
This token is generated in Coscine and is intended for machine use of Coscine. Once users have entered their token on the website, they are redirected to the project page. This project page contains all projects and sub-projects to which the users belong. The user can then select the project and resource to which they wish to upload their FACS files. The website will load a variety of file types if they match the FACS metadata format and the required fields are filled in. If changes are needed after the file is uploaded to Coscine, users can make them in Coscine.
This upload website is still under development, with the goal of extending its use to more types of files so that other Python scripts can be invoked that extract metadata from different files. To enable this, it is planned that a drop-down list will appear on the homepage where users can specify the type of file they want to upload to Coscine. Users will then be directed to the appropriate resources that contain the application profile specific to the file type. The appropriate resources are then listed.
Once this site is complete it will run a variety of code in the background to extract metadata and store files easily in Coscine. Thus, limiting the burden on the researchers and making it effortless to manage research data FAIRly.
Learn more
You don’t want to miss any news about Coscine? Then subscribe to our mailing list and visit us on our website. You can find comprehensive instructions on how to use Coscine in our documentation.
Do you have questions or feedback? Then write a message to the IT-ServiceDesk. We are looking forward to your message!
________
Responsible for the content of this article are Catherine Gonzalez and Arlinda Ujkani.





Leave a Reply
You must be logged in to post a comment.